Basic Nat Functions
Registering a light-level tracing
First, load in the light-level tracing. This tracing was generated using the Simple Neurite Tracer plugin in Fiji and saved as a .swc file
neuronTracing <- read.neuron('data/Namiki_tracing_L_2.swc',class="neuron")
open3d(userMatrix=rotationMatrix,windowRect=windowRect,zoom=0.7)
plot3d(neuronTracing,lwd=3,col='black',WithNodes=FALSE)You must enable Javascript to view this page properly.
Next, load in the regisration text file generated through CMTK to register your neuron into the desired template brain space. Apply this transformation to your tracing using xform
reg <- as.cmtkreg('data/Namiki_Registration_JFRC2013.txt')
registeredNeuron <- xform(neuronTracing, reg)
#Plot neuron along with registered brain template
open3d(userMatrix=rotationMatrix,windowRect=windowRect,zoom=0.7)
plot3d(registeredNeuron,lwd=3,col='black',WithNodes=FALSE)
plot3d(JFRC2013)
#save the registered neuron to a .swc file
write.neuron(registeredNeuron,'data/registeredNeuron_JFRC2013',format='swc')You must enable Javascript to view this page properly.
Transform a neuron between template brains
Once a neuron is registered to a standard template brain, it can be transformed into another template brain space. A graph of all available tempalte brains and their transformations are shown below: 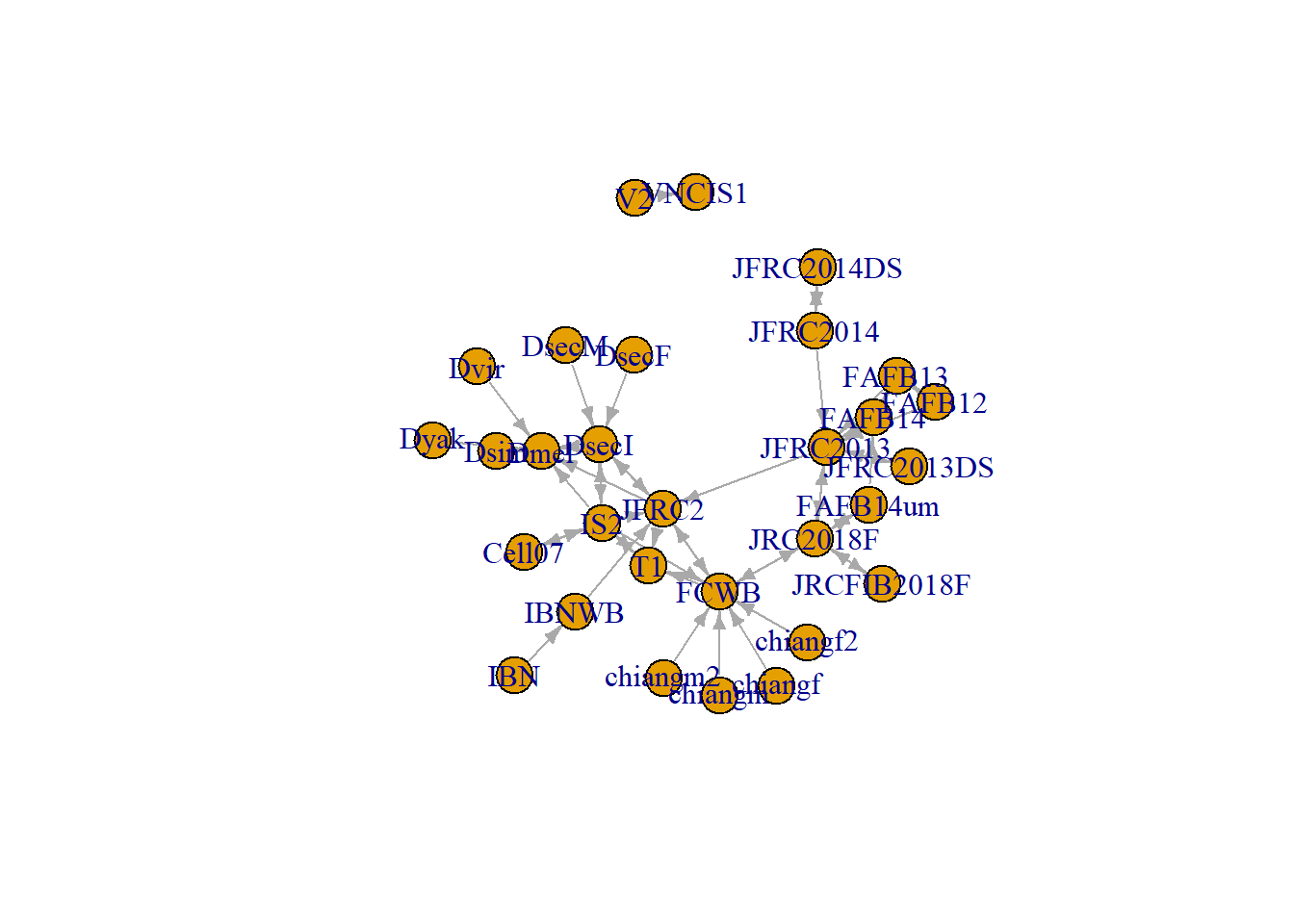
The function xform_brain is used to complete this transformation
#transform neuron from JFRC2013 template brain space to JFRC2 template brain space
#sample=template brain that the neuron is currently registered to
#reference= template brain that the neuron will be transformed into
neuron_JFRC2=xform_brain(registeredNeuron,sample=JFRC2013,reference=JFRC2)
#plot the neuron with its new template brain
open3d(userMatrix=rotationMatrix,windowRect=windowRect,zoom=0.7)
plot3d(neuron_JFRC2,lwd=3,col='black',WithNodes=FALSE)
plot3d(JFRC2)You must enable Javascript to view this page properly.
Working with FlyCircuit data
Through the flycircuit package, you can work with FlyCircuit skeletons in R. These skeletons can be manipulated like any other neuron object using the suite of natverse tools.
To read in a flycircuit neuron, query the flycircuit database with your desired flycircuit id. (Note: the flycircuit skeletons fetched this way are registered to the JFRC2 template brain )
flycircuit_id <- flycircuit::fc_idid('fru-M-100201')
query <- sprintf(
"http://www.virtualflybrain.org/data/VFB/i/%04d/%04d/volume.swc",
flycircuit_id %/% 1e4,
flycircuit_id %% 1e4
)
flycircuit_neuron=read.neurons(query, neuronnames = flycircuit::fc_neuron('fru-F-000100'))
#plot flycircuit neuron
open3d(userMatrix=rotationMatrix,windowRect=windowRect,zoom=0.7)
plot3d(flycircuit_neuron,lwd=3,col='black',WithNodes=FALSE)
plot3d(JFRC2)You must enable Javascript to view this page properly.